Our Research
Opine Metallophore Project
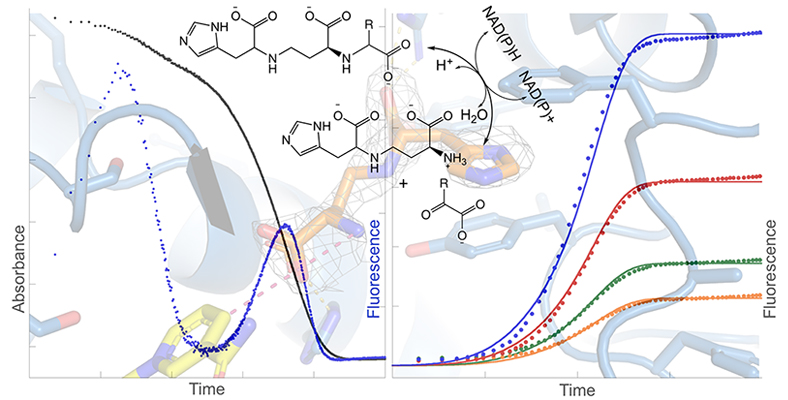
Nicotianamine (NA) is a metabolite synthesized by plants to maintain homeostasis of metal ion micronutrients such as iron, nickel and zinc. In certain plants it has an additional function of serving as a precursor of phytosiderophores that perform extracellular iron scavenging. More recently, opine-type metallophores homologous to NA were found in bacteria, especially in infectious human pathogens such as Staphylococcus aureus, Pseudomonas aeruginosa, and Yersinia pestis. In bacteria, opine metallophores are produced by two enzymes, a nicotianamine synthase (NAS) and an opine dehydrogenase (ODH). The canonical nicotianamine synthases from plants are aminoalkyltransferases that biosynthesize a siderophore nicotianamine, processively linking together aminobutyrate moieties derived from S-adenosylmethionine (SAM). To date, NAS enzymes from pathogenic bacteria have been shown to catalyze the linkage of an aminobutyrate from SAM to an amino acid. ODH enzymes catalyze the addition of an α-keto acid to the product of the NAS reaction. We use structural biology and mechanistic enzymology to determine how these enzymes catalyze their particular reactions. One key question we are currently answering is what makes the plant NAS processive, whereas the bacterial enzymes are not. This basic science research lays the necessary foundation for future novel antimicrobial drug design efforts.
Riboflavin Project
Riboflavin is required by all living organisms and is synthesized from one GTP and two ribulose 5-phosphate molecules using five unique enzymes. The chemistry at work in this pathway is specialized and involves enzymes that have no paralogs. Despite the unparalleled chemistry and the ubiquitous requirement for riboflavin, compelling evidence for the chemical mechanisms of the enzymes in this biosynthetic pathway has not been presented. We are working to fill that gap in knowledge. The results will provide key information for other researchers who work toward the generation of novel antimicrobials in the fight against multidrug resistant bacterial pathogens, and for parasitic pathogens dependent on bacterial symbionts to generate the vitamin riboflavin.

Collaborations
- Collaboration with Dr. Graham Moran at Loyola University Chicago
- Many, including the above projects
- Collaboration with Dr. Thomas Meek at Texas A&M University, Department of Biochemistry and Biophysics
- Crystallographic structure determination of several cysteine proteases including the protease of SARS-Co-V2 (COVID-19)
- Crystallographic structure determination of purine phosphoribosyltransferases, in particular those pertaining to malaria, trypanonomiasis, and Chagas’s disease
- Collaboration with Dr. Aron Fenton and Dr. Liskin Swint-Kruse at the University of Kansas Medical Center
- Structure studies of pyruvate kinase, aldolase, phosphofructokinase, and pyruvate carboxylase
- Collaboration with Dr. Karl Klose at The University of Texas at San Antonio, Department of Molecular Microbiology and Immunology
- Structural studies of flagellar regulatory protein B (FlrB)

