Current Research
Mechanisms regulating the specification, maintenance and loss of SSC fate
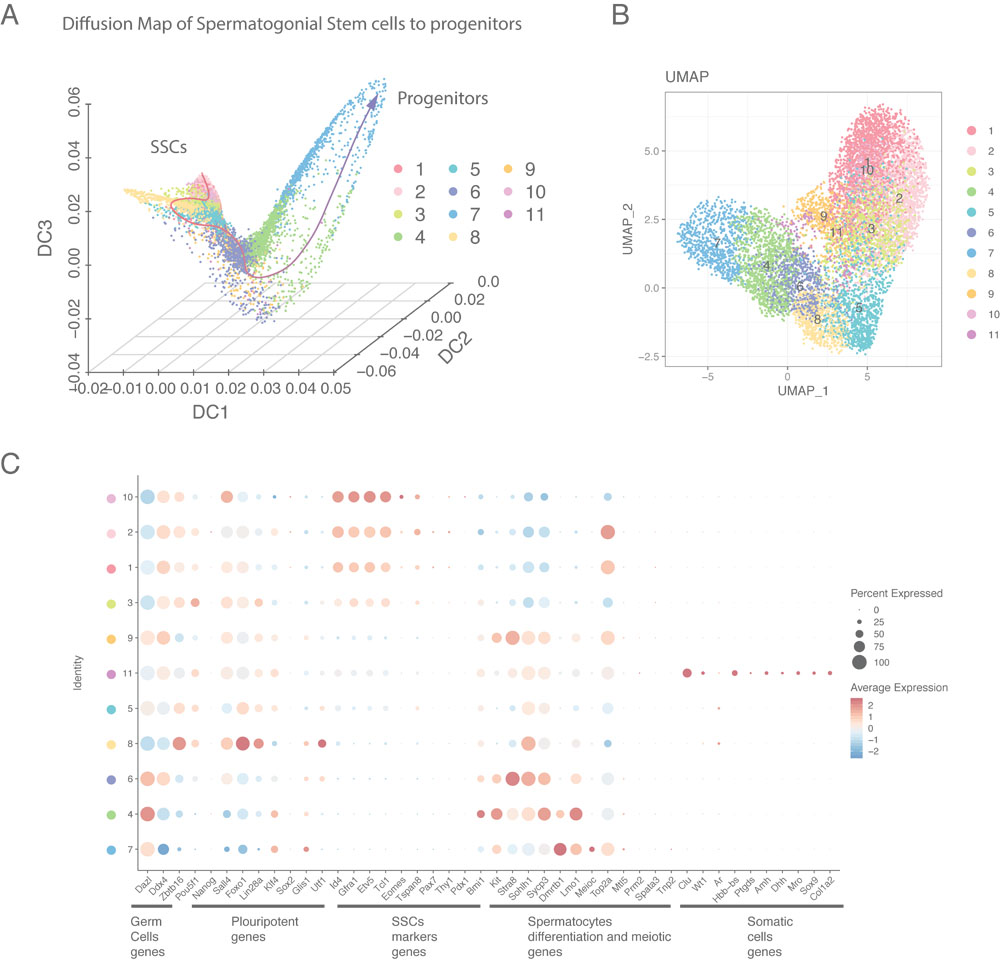 Spermatogonial stem cells (SSCs) sustain the seminiferous epithelium in the testis and maintain steady-state spermatogenesis by a balance between self-renewal and production of progenitor spermatogonia that enter the spermatogenic differentiation pathway. In collaboration with Drs. Brian Hermann at The University of Texas at San Antonio, Christopher Geyer at East Carolina University, and John Oatley at Washington State University, we are studying differential gene expression and related epigenetic programming associated with each of these spermatogonial subtypes. We have characterized subtype-specific gene expression by both bulk and single-cell RNA-seq, and associated epigenetic programming by multiparametric integrative epigenomic profiling including analyses of genome-wide patterns of DNA methylation, chromatin accessibility and six different histone modification patterns (H3K4me1/2/3, H3K9me3, H3K27me3, and H3K27ac). Our results suggest that SSCs and progenitors are distinct spermatogonial subtypes differentially programmed to either self-renew and maintain regenerative capacity as SSCs, or lose regenerative capacity as progenitors and initiate lineage commitment associated with spermatogenic differentiation.
Spermatogonial stem cells (SSCs) sustain the seminiferous epithelium in the testis and maintain steady-state spermatogenesis by a balance between self-renewal and production of progenitor spermatogonia that enter the spermatogenic differentiation pathway. In collaboration with Drs. Brian Hermann at The University of Texas at San Antonio, Christopher Geyer at East Carolina University, and John Oatley at Washington State University, we are studying differential gene expression and related epigenetic programming associated with each of these spermatogonial subtypes. We have characterized subtype-specific gene expression by both bulk and single-cell RNA-seq, and associated epigenetic programming by multiparametric integrative epigenomic profiling including analyses of genome-wide patterns of DNA methylation, chromatin accessibility and six different histone modification patterns (H3K4me1/2/3, H3K9me3, H3K27me3, and H3K27ac). Our results suggest that SSCs and progenitors are distinct spermatogonial subtypes differentially programmed to either self-renew and maintain regenerative capacity as SSCs, or lose regenerative capacity as progenitors and initiate lineage commitment associated with spermatogenic differentiation.
- Cheng K, Chen I-C, Hale B, Hermann BP, Geyer CB, Oatley JM, McCarrey JR. Unique epigenetic programming distinguishes regenerative spermatogonial stem cells in the developing mouse testis. Nat Comms, in revision. bioRxiv 674457. doi:10.1101/674457
- Hermann BP, Singh A, Cheng K, Mutoji KN, Roa-de-la-Cruz L, Gilersleeve H, Chen I-C, Mayo M, Westernstroer B, Law NC, Oatley MJ, Velte EK, Niedenberger BA, Fritze D, Silber S, Geyer CB, Oatley JM, McCarrey JR. (2018) The mammalian spermatogenesis single cell transcriptome – from spermatogonial stem cells to spermatids. Cell Reports 25:1650-1667. doi:10.1016/j.celrep.2018.10.026 | PMID:30404016.
- McCarrey JR. (2017) Transition of prenatal prospermatogonia to postnatal spermatogonia. In: The Biology of Mammalian Spermatogonia. J. Oatley, M. Griswold, eds. Springer Publishing Co. New York, NY.
- Mutoji K, Singh A, Nguyen T, Gildersleeve H, Kaucher AV, Oatley MJ, Oatley JM, Velte EK, Geyer CB, Chen K, McCarrey JR, Hermann BP. (2016) TSPAN8 expression distinguishes spermatogonial stem cells in the prepubertal testis. Biol Reprod Dec;95(6):117.
- Hermann BP, Mutoji K, Velte EK, Ko D, Oatley JM, Geyer CB, McCarrey JR. (2015) Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod 92:54. doi:10.1095/biolreprod.114.125757
Potential for Assisted Reproductive Technologies (ART) to disrupt epigenetic programming or genetic integrity
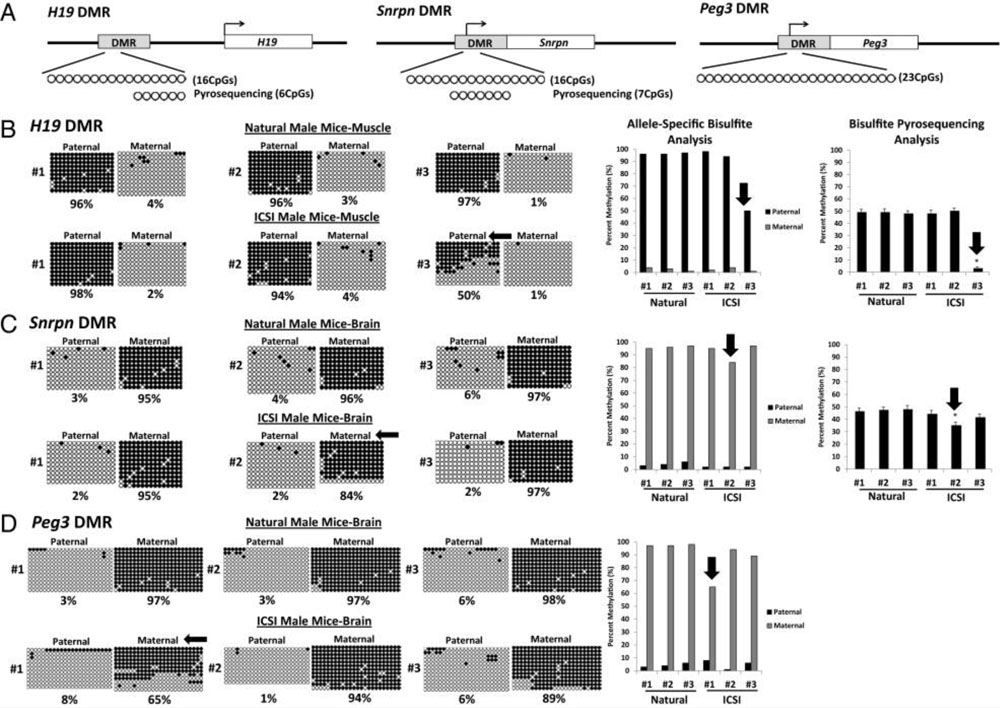 Previous reports from our lab and others have indicated that methods of ART may predispose epimutations in the ensuing offspring – particularly in imprinted genes. In collaboration with Drs. Alex Meissner from the Max Planck Institute in Berlin and Yukiko Yamazaki from the University of Hawaii in Honolulu, we have conducted a comprehensive analysis of gene expression and epigenetic programming in mice generated by either intracytoplasmic sperm injection (ICSI) or natural reproduction. Initial studies were focused on epigenetic programming at specific imprinted loci in mice produced by ICSI or natural mating. More recent studies have included genome-wide assessments of gene expression and DNA methylation patterns in mice produced by ICSI or natural mating. Reassuringly, our results indicate that genome-wide ART methods induce very few epimutations or aberrant gene expression patterns.
Previous reports from our lab and others have indicated that methods of ART may predispose epimutations in the ensuing offspring – particularly in imprinted genes. In collaboration with Drs. Alex Meissner from the Max Planck Institute in Berlin and Yukiko Yamazaki from the University of Hawaii in Honolulu, we have conducted a comprehensive analysis of gene expression and epigenetic programming in mice generated by either intracytoplasmic sperm injection (ICSI) or natural reproduction. Initial studies were focused on epigenetic programming at specific imprinted loci in mice produced by ICSI or natural mating. More recent studies have included genome-wide assessments of gene expression and DNA methylation patterns in mice produced by ICSI or natural mating. Reassuringly, our results indicate that genome-wide ART methods induce very few epimutations or aberrant gene expression patterns.
- de Waal E, Yamazaki Y, Ingale P, Bartolomei MS, Yanagimachi R, McCarrey JR. (2012) Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci (USA), 109: 4163-4168. doi:10.1073/pnas.1201990109
- de Waal E, Yamazaki Y, Ingale P, Bartolomei M, Yanagimachi R, McCarrey JR (2012) Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum Molec Genet 21: 4460-4472. doi:10.1093/hmg/dds287
- de Waal E, McCarrey JR. (2010) Effects of exogenous endocrine stimulation on epigenetic programming of the female germline genome. Animal Reprod, 7:154-164.
- Murphey P, Yamazaki Y, McMahan CA, Walter CA, Yanagimachi R, McCarrey JR. (2009) Epigenetic regulation of genetic integrity is reprogrammed during cloning. Proc Natl Acad Sci (USA) 106:4731-4735. doi:10.1073/pnas.0900687106
- McCarrey JR. (2009) Maintenance of genetic integrity during natural and assisted reproduction. Reproductive BioMedicine Online 18 (Suppl 2):51-55. doi:10.1016/s1472-6483(10)60449-x
- Caperton L, Murphey P, Yamazaki Y, McMahan CA, Walter CA, Yanagimachi R, McCarrey JR. (2007) Assisted reproductive technologies do not alter mutation frequency or spectrum. Proc. Natl. Acad. Sci. (USA) 104: 5085-5090. doi:10.1073/pnas.0611642104
Effects of male lifestyle choices on the sperm epigenome and intergenerational epigenetic inheritance of paternal epimutations
 We recently initiated a project that is part of a new Center for Male Reproductive Epigenomics which is one of the NICHD-sponsored National Centers for Translational Research in Reproduction and Infertility (NCTRI) and is headed by Dr. Wei Yan at UCLA and includes additional UCLA collaborators – Drs. Ron Swerdloff, Christina Wang and Harry Rossiter. The McCarrey lab project involves characterization of epigenetic profiling of the sperm epigenome in mice and men who have consumed either a healthy or a high fat diet, with or without an associated exercise regime. The male mice will be allowed to generate F1 offspring to determine if paternal diet and exercise can impact the male's offspring. We will also assess the extent to which any epimutations in the sperm epigenome associated with consumption of a high fat diet with no exercise can be reversed by transition to consumption of a healthy diet with regular exercise. This center also organizes a community engagement and education core designed to increase community awareness regarding the extent to which a man's lifestyle (diet and exercise) prior to conception may impact the health of his children.
We recently initiated a project that is part of a new Center for Male Reproductive Epigenomics which is one of the NICHD-sponsored National Centers for Translational Research in Reproduction and Infertility (NCTRI) and is headed by Dr. Wei Yan at UCLA and includes additional UCLA collaborators – Drs. Ron Swerdloff, Christina Wang and Harry Rossiter. The McCarrey lab project involves characterization of epigenetic profiling of the sperm epigenome in mice and men who have consumed either a healthy or a high fat diet, with or without an associated exercise regime. The male mice will be allowed to generate F1 offspring to determine if paternal diet and exercise can impact the male's offspring. We will also assess the extent to which any epimutations in the sperm epigenome associated with consumption of a high fat diet with no exercise can be reversed by transition to consumption of a healthy diet with regular exercise. This center also organizes a community engagement and education core designed to increase community awareness regarding the extent to which a man's lifestyle (diet and exercise) prior to conception may impact the health of his children.
• Healthy Lifestyle • Healthy Sperm • Healthy Children
Epigenetic reprogramming in a dish - an in vitro model of transgenerational epigenetic inheritance
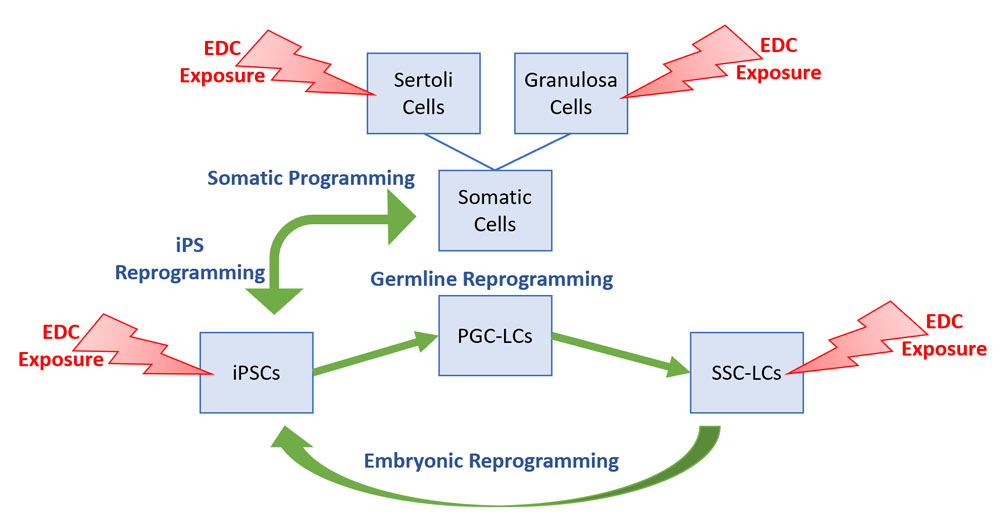 Disrupting chemicals or other environmental effects can induce epimutations that can predispose development of disease states, and can be transmitted via inter- or transgenerational epigenetic inheritance to subsequent generations. These findings are the result of large-scale studies conducted in animals (e.g. mice or rats) that, while informative, afford low resolution of the underlying mechanism(s) and are time-, cost-, labor-, and animal-intense. We are developing a cell culture system in which the normal cycle of embryonic and germline epigenetic reprogramming can be recapitulated in vitro to facilitate high-resolution, time-, cost-, labor-, and animal-saving studies at the cellular and molecular levels to discern the manner in which exposure to environmental disruptors leads to the initial induction of epimutations, and how these epimutations subsequently escape, and/or circumvent the potentially corrective effects of epigenetic reprogramming such that they are transmitted to subsequent generations.
Disrupting chemicals or other environmental effects can induce epimutations that can predispose development of disease states, and can be transmitted via inter- or transgenerational epigenetic inheritance to subsequent generations. These findings are the result of large-scale studies conducted in animals (e.g. mice or rats) that, while informative, afford low resolution of the underlying mechanism(s) and are time-, cost-, labor-, and animal-intense. We are developing a cell culture system in which the normal cycle of embryonic and germline epigenetic reprogramming can be recapitulated in vitro to facilitate high-resolution, time-, cost-, labor-, and animal-saving studies at the cellular and molecular levels to discern the manner in which exposure to environmental disruptors leads to the initial induction of epimutations, and how these epimutations subsequently escape, and/or circumvent the potentially corrective effects of epigenetic reprogramming such that they are transmitted to subsequent generations.
- Lehle JD, McCarrey JR. Differential susceptibility to endocrine disruptor-induced epimutagenesis. Environmental Epigenetics, 2020 Dec 8; 6(1):dvaa016. doi:10.1093/eep/dvaa016.
Differential epigenetic reprogramming potential among pluripotent, germ and somatic cell types
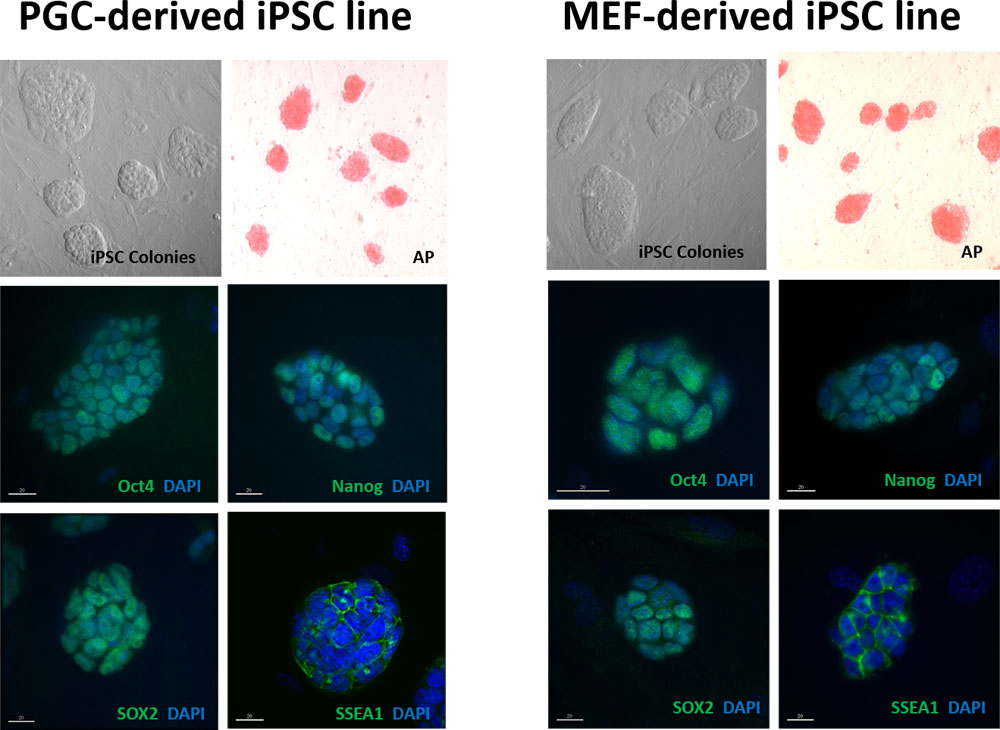 When different cell types are reprogrammed into induced pluripotent cells (iPSCs), they can retain residual epigenetic memory of their previous differentiated state. This residual epigenetic memory can influence the potential to fully reprogram and ultimately re-differentiate specific cell types for use in research or therapeutic applications. We are pursuing research to determine the extent to which residual epigenetic programming from source cell types limits normal epigenetic reprogramming in association with each of the following transitions in cell fate in vitro: 1) somatic to pluripotent, 2) pluripotent to early germ line, 3) early germ line to later germ line, and 4) germ line back to pluripotent. Our initial goal is to establish an in vitro system in which transitions among pluripotent, germ or somatic cell states can be induced and monitored, so that we can then study in vitro the epigenetic programming or reprogramming that normally accompanies similar transitions during development in vivo.
When different cell types are reprogrammed into induced pluripotent cells (iPSCs), they can retain residual epigenetic memory of their previous differentiated state. This residual epigenetic memory can influence the potential to fully reprogram and ultimately re-differentiate specific cell types for use in research or therapeutic applications. We are pursuing research to determine the extent to which residual epigenetic programming from source cell types limits normal epigenetic reprogramming in association with each of the following transitions in cell fate in vitro: 1) somatic to pluripotent, 2) pluripotent to early germ line, 3) early germ line to later germ line, and 4) germ line back to pluripotent. Our initial goal is to establish an in vitro system in which transitions among pluripotent, germ or somatic cell states can be induced and monitored, so that we can then study in vitro the epigenetic programming or reprogramming that normally accompanies similar transitions during development in vivo.
Developing the baboon as a model system for cell-based therapies
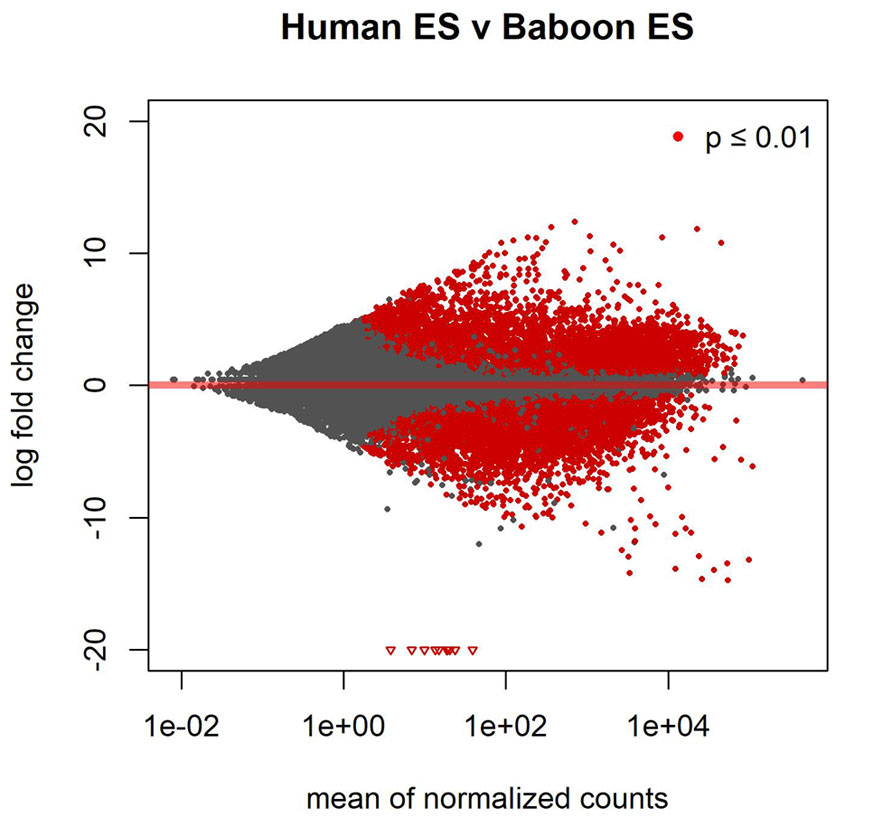
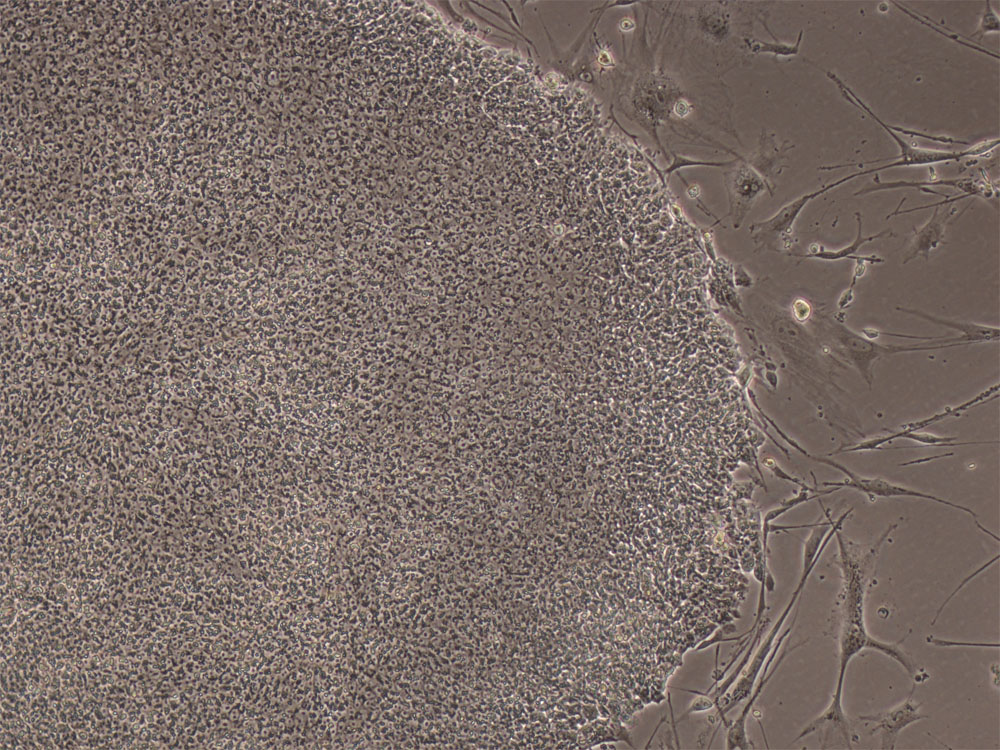 Stem cell-based therapies have the potential to dramatically improve the treatment and prognosis of patients suffering from a variety of debilitating or degenerative diseases, or from injury or battlefield trauma by restoring cell or tissue function. Yet, the advancement of stem cell-based therapies requires optimization of efficacy and safety of these novel methodologies, and this requires preclinical validation in model organisms prior to transition into the clinic. Nonhuman primate (NHP) species provide the most accurate recapitulation of the human condition for use in preclinical testing. The baboon (Papio anubis) represents the most attractive NHP species for assessments of stem cell therapies because they are similar to humans in size and anatomy, as well as in development, physiology and neurological functions. In addition, baboons possess an immunogenetic system that is more like that of humans than that found in other NHPs. We have validated baboon ESCs and iPSCs using gene expression and epigenetic profiling analyses, and we are optimizing the use of a "scorecard" method to assess pluripotency and differentiation potential of baboon stem cell lines. Our long-term goal is to establish the baboon as a model NHP species for testing and optimizing the efficacy and safety of stem cell-based therapeutic approaches.
Stem cell-based therapies have the potential to dramatically improve the treatment and prognosis of patients suffering from a variety of debilitating or degenerative diseases, or from injury or battlefield trauma by restoring cell or tissue function. Yet, the advancement of stem cell-based therapies requires optimization of efficacy and safety of these novel methodologies, and this requires preclinical validation in model organisms prior to transition into the clinic. Nonhuman primate (NHP) species provide the most accurate recapitulation of the human condition for use in preclinical testing. The baboon (Papio anubis) represents the most attractive NHP species for assessments of stem cell therapies because they are similar to humans in size and anatomy, as well as in development, physiology and neurological functions. In addition, baboons possess an immunogenetic system that is more like that of humans than that found in other NHPs. We have validated baboon ESCs and iPSCs using gene expression and epigenetic profiling analyses, and we are optimizing the use of a "scorecard" method to assess pluripotency and differentiation potential of baboon stem cell lines. Our long-term goal is to establish the baboon as a model NHP species for testing and optimizing the efficacy and safety of stem cell-based therapeutic approaches.
- Mahlke MA, Cheng K, Li B, Chaudhari S, Navara CS, McCarrey JR. Validation of baboon pluripotent cells as a model for translational stem cell research. Stem Cell Res. 2021 Nov 12:57:102598. doi:10.1016/j.scr.2021.102598
- Navara CS, Chaudhari S, McCarrey JR. (2018) Optimization of culture conditions for the derivation and propagation of baboon (Papio Anubis) induced pluripotent stem cells. PLoS One 13(3): e0193195. eCollection 2018. doi:10.1371/journal.pone.0193195
- Grow DA, McCarrey JR, Navara CS. (2016) Advantages of nonhuman primates as preclinical models for evaluating stem cell-based therapies for Parkinson's Disease. Stem Cell Res Sep;17(2):352-366. doi:10.1016/j.scr.2016.08.013
- Grow DA, Simmons D, Gomez JA, Wanat MJ, McCarrey JR, Paladini CA, Navara CS. (2016) Differentiation and characterization of dopaminergic neurons from baboon pluripotent stem cells. Stem Cells – Translational Medicine Sep;5(9):1133-44. doi:10.5966/sctm.2015-0073
- Navara C, Hornecker J, Grow D, Chaudhari S, Hornsby P, Ichida J, Eggan K, McCarrey JR. (2013) Derivation of induced pluripotent stem cells from the baboon: A nonhuman primate model for preclinical testing of stem cell therapies. Cell Reprogram 15: 495-502. doi:10.1089/cell.2012.0093
Past Research
 For several decades, research in the McCarrey laboratory is centered on mammalian germ cells and stem cells. We have made use of several experimental models, including the mouse, baboon, and opossum. Dr. McCarrey discovered the first example of a functional, germ-cell-specific retroposon in the human genome. He has published several papers on mechanisms that regulate germ-cell-specific gene expression in mammals.
For several decades, research in the McCarrey laboratory is centered on mammalian germ cells and stem cells. We have made use of several experimental models, including the mouse, baboon, and opossum. Dr. McCarrey discovered the first example of a functional, germ-cell-specific retroposon in the human genome. He has published several papers on mechanisms that regulate germ-cell-specific gene expression in mammals.
 Dr. McCarrey has also published several papers on mechanisms of epigenetic programming that function during embryogenesis, germ cell development and gametogenesis, as well as papers focused on mechanisms governing X-chromosome activity or inactivity in germ cells and early embryos, the effects of cloning and assisted reproductive technologies on genetic integrity, epigenetic programming in gametes and embryos, the relationship between pluripotency and maintenance of genetic integrity, and the development of nonhuman primate model systems for studies of stem cell research and regenerative medicine. Finally, he also maintains an active interest in the evolution of genetic and epigenetic mechanisms of gene regulation in mammals.
Dr. McCarrey has also published several papers on mechanisms of epigenetic programming that function during embryogenesis, germ cell development and gametogenesis, as well as papers focused on mechanisms governing X-chromosome activity or inactivity in germ cells and early embryos, the effects of cloning and assisted reproductive technologies on genetic integrity, epigenetic programming in gametes and embryos, the relationship between pluripotency and maintenance of genetic integrity, and the development of nonhuman primate model systems for studies of stem cell research and regenerative medicine. Finally, he also maintains an active interest in the evolution of genetic and epigenetic mechanisms of gene regulation in mammals.
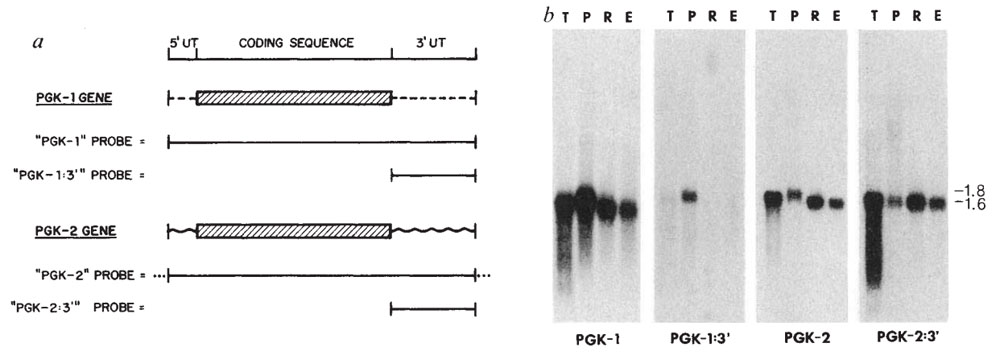
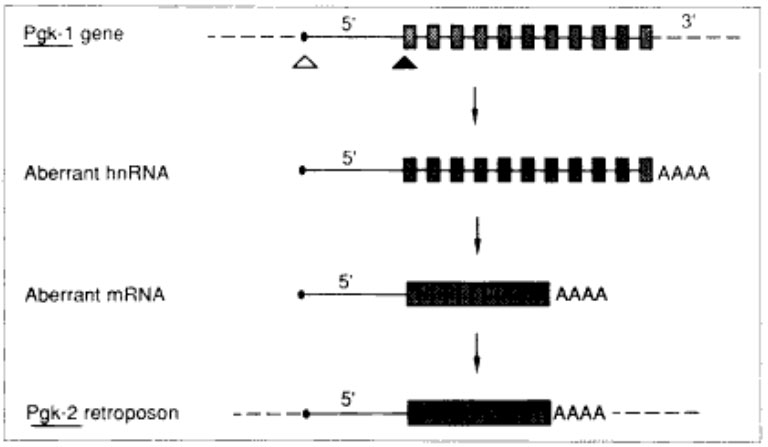 In 1987, Dr. McCarrey reported the first example of a functional retroposon in the human genome – the autosomal PGK2 gene. Evidence suggests that this intronless member of the PGK gene family arose as a reverse-transcribed copy of an mRNA from the intron-containing, X-linked PGK1 gene. This is an example of Susumu Ohno's theory of "Evolution by Gene Duplication." Following its origin, the PGK2 retrogene evolved from a ubiquitously expressed housekeeping gene to a tightly regulated tissue-specific gene expressed only during spermatogenesis in male eutherian mammals and encoding a protein that functions uniquely in sperm.
In 1987, Dr. McCarrey reported the first example of a functional retroposon in the human genome – the autosomal PGK2 gene. Evidence suggests that this intronless member of the PGK gene family arose as a reverse-transcribed copy of an mRNA from the intron-containing, X-linked PGK1 gene. This is an example of Susumu Ohno's theory of "Evolution by Gene Duplication." Following its origin, the PGK2 retrogene evolved from a ubiquitously expressed housekeeping gene to a tightly regulated tissue-specific gene expressed only during spermatogenesis in male eutherian mammals and encoding a protein that functions uniquely in sperm.
- McCarrey, J.R. (1994) Evolution of tissue-specific gene expression in mammals: A new gene is formed and refined. Bioscience 44: 20-27.
- McCarrey, J.R. (1990) Molecular evolution of the Pgk-2 retroposon. Nucleic Acids Res. 18: 949-955.
- McCarrey, J.R. (1987) Nucleotide sequence of the promoter region of a tissue-specific retroposon: Comparison with its housekeeping progenitor. GENE 61: 291-298.
- McCarrey, J.R. and Thomas, K. (1987) Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature 326: 501-505.
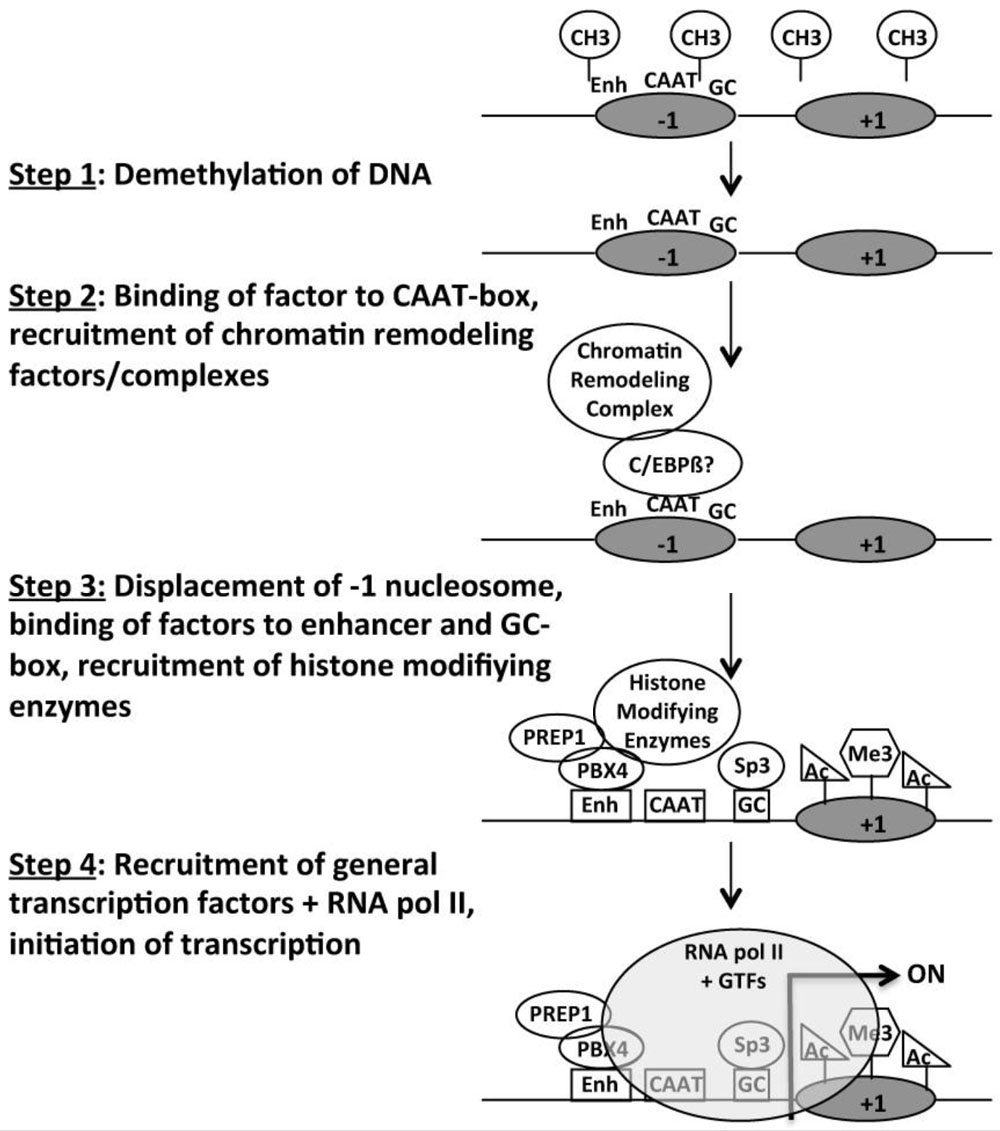 Studies of the spermatogenesis-specific Pgk2 gene in the mouse during the 1980s and 90s and early 2000s yielded novel information about the manner in which a spermatogenesis-specific gene promoter functions, including protein-DNA interactions, the identification of key transcription factors regulating spermatogenesis-specific expression, and changes in DNA methylation, histone modifications, nucleosome positioning, and chromatin structure associated with transcriptional activation of gene expression in spermatogenic cells.
Studies of the spermatogenesis-specific Pgk2 gene in the mouse during the 1980s and 90s and early 2000s yielded novel information about the manner in which a spermatogenesis-specific gene promoter functions, including protein-DNA interactions, the identification of key transcription factors regulating spermatogenesis-specific expression, and changes in DNA methylation, histone modifications, nucleosome positioning, and chromatin structure associated with transcriptional activation of gene expression in spermatogenic cells.
- Yang Z, Yoshioka H, McCarrey JR. (2013) Sequence-specific promoter elements regulate temporal-specific changes in chromatin required for testis-specific activation of the Pgk2 gene. Reproduction 146: 501-516.
- Yoshioka H, Geyer CB, Hornecker JL, Patel K, McCarrey JR. (2007) Analysis in vivo of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during spermatogenesis. Mol Cell Biol 27: 7871-7885.
- McCarrey JR, Geyer CB, Yoshioka Y. (2005) Epigenetic regulation of testis-specific gene expression. In: Testicular Cell Dynamics and Endocrine Signaling, vol. 1061 of the Annals of the New York Academy of Sciences. MP Hardy and MD Griswold (eds). Vol. 1061, pp 226-242.
- Shima JE, McLean DJ, McCarrey JR, Griswold, MD. (2004) The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 71: 319-330.
- Geyer CB, Kiefer CM, Yang TP, McCarrey JR. (2004) Ontogeny of a demethylation domain and its relationship to activation of tissue-specific transcription. Biol. Reprod. 71: 837-844.
- McCarrey, J.R. Epigenetic mechanisms regulating gene expression. (2003) In: Introduction to Bioinformatics, Chapter 6. S.A. Krawetz & D.D. Womble, eds. Humana Press Inc. Totowa, NJ. pp 123-150.
- Zhang, L.P., Stroud, J., Eddy, C.A., Walter, C.A. and McCarrey, J.R. (1999) Multiple elements influence transcriptional regulation from the human testis-specific PGK2 promoter in transgenic mice. Biol. Reprod. 60: 1329-1337.
- McCarrey, J.R. (1998) Spermatogenesis as a model system for developmental analysis of regulatory mechanisms associated with tissue-specific gene expression. Seminars in Cell & Devel. Biol. 9: 459-466.
- Zhang, L.P., Stroud, J.C., Walter, C.A., Adrian, G.S., and McCarrey, J.R. (1998) A gene-specific promoter in transgenic mice directs testis-specific demethylation prior to transcriptional activation in vivo. Biol. Reprod. 59: 284-292.
- Kumari, M., Stroud, J.C., Anji, A., and McCarrey, J.R. (1996) Differential Appearance of DNase I-hypersensitive sites correlates with differential transcription of Pgk genes during spermatogenesis in the mouse. J. Biol. Chem. 271: 14390-14397.
- McCarrey, J.R. (1996) Regulatory mechanisms distinguishing ubiquitous and testis-specific expression of the mammalian Pgk-1 and Pgk-2 genes. 8th International Congress on Isozymes In: Gene Families: Structure, Function, Genetics and Evolution. R.S. Holmes & H.A. Lim (eds). World Scientific. London. pp 159-164.
- Berg, W.M., Gebara, M.M. and McCarrey, J.R. (1993) Sp1 is required for initiation and stimulation of transcription from the TATA-less PGK-2 promoter. Life Sci. Adv. - Mol. Biol. 12: 85-91.
- Urven, L.E., Weng, D.E., Schumaker, A.L., Gearhart, J.D., and McCarrey, J.R. (1993) Differential gene expression in fetal mouse germ cells. Biol. Reprod. 48: 564-574.
- McCarrey, J.R. and Dilworth, D.D. (1992) Expression of Xist in mouse germ cells correlates with X-chromosome inactivation. Nature Genetics 2: 200-203.
- McCarrey, J.R., Berg, W.M., Paragioudakis, S.J., Zhang, P.L., Dilworth, D.D., Arnold, B.L., and Rossi, J.J. (1992). Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev. Biol. 154: 160-168.
- Gebara, M.M. and McCarrey, J.R. (1992) Protein-DNA interactions associated with the onset of testis-specific expression of the Pgk-2 gene. Mol. Cell. Biol. 12: 1422-1431.
- Robinson, M.O., McCarrey, J.R., and Simon, M.I. (1989) Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proc. Nat'l. Acad. Sci. (USA) 86: 8437-8441.
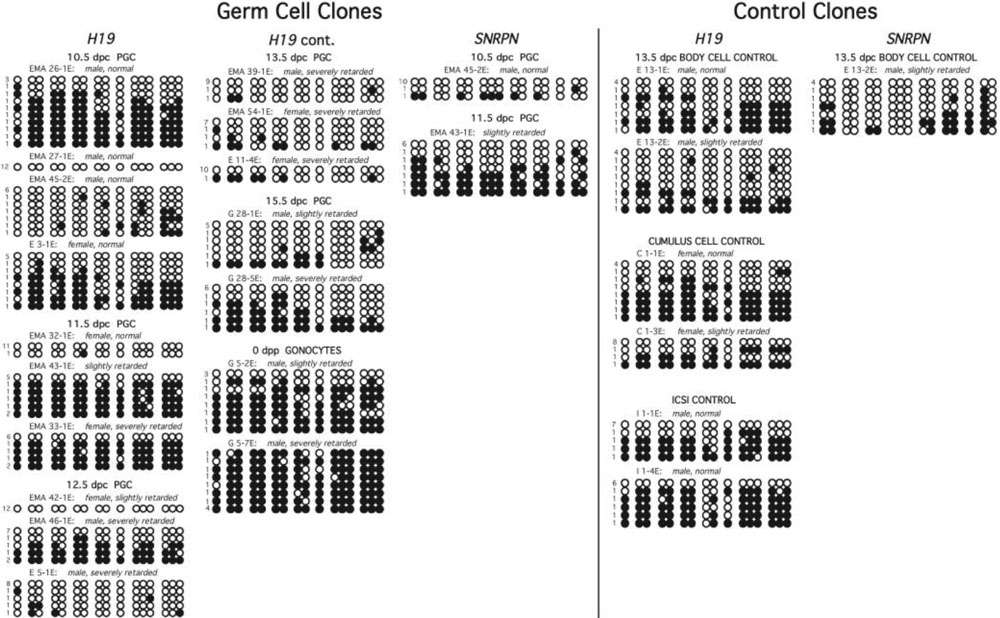 Throughout the 1990s – 2000s, Dr. McCarrey was engaged in collaborative studies with Dr. Howard Cedar at the Hebrew University in Jerusalem describing epigenetic reprogramming during germline development, including methylation patterns of testis-specific genes, reprogramming of DNA methylation patterns genome-wide, germline-specific reprogramming of imprinted genes. Studies regarding normal and disrupted epigenetic programming in the germ line have continued in the McCarrey lab and are currently ongoing.
Throughout the 1990s – 2000s, Dr. McCarrey was engaged in collaborative studies with Dr. Howard Cedar at the Hebrew University in Jerusalem describing epigenetic reprogramming during germline development, including methylation patterns of testis-specific genes, reprogramming of DNA methylation patterns genome-wide, germline-specific reprogramming of imprinted genes. Studies regarding normal and disrupted epigenetic programming in the germ line have continued in the McCarrey lab and are currently ongoing.
- Yamazaki, Y., Lee, S.S., Marh, J., McCarrey, J.R., Mann, M.R.W., Yanagimachi, R., and Bartolomei, M.S. (2003) Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc. Natl. Acad. Sci. 100: 12207-12212.
- Klaus, A., McCarrey, J.R., Farkas, A.and Ward, W.S. (2001) Changes in DNA loop domain structure during spermatogenesis and embryogenesis. Biology of Reproduction 64: 1297-1306.
- Simon, I., Tanzen, T., Rubinoff, B., McCarrey, J. and Cedar, H. (1999) Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401: 929-932.
- Ariel, M., Robinson, E., McCarrey, J. and Cedar, H. (1995) Gamete specific methylation correlates with imprinting of the murine Xist gene. Nature Genetics 9: 312-315.
- Ariel, M., Cedar, H., and McCarrey, J.R. (1994) Developmental changes in Pgk-2 gene methylation during spermatogenesis in the mouse: Reprogramming occurs in epididymal spermatozoa. Nature Genetics 7: 59-63.
- Brandeis, M., Kafri, T., Ariel, M., Chaillet, J.R., McCarrey, J.R., Razin, A., and Cedar, H. (1993) The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 12: 3669-3677.
- Kafri, T., Ariel, M., Shemer, R., Urven, L., McCarrey, J.R., Cedar, H. and Razin, A. (1992) Dynamics of gene specific DNA methylation in the mouse embryo and germ line. Genes & Development 6: 705-714.
- Ariel, M., McCarrey, J., and Cedar, H. (1991) Methylation patterns of testis-specific genes. Proc. Nat'l. Acad. Sci. (USA) 88: 2317-2321.
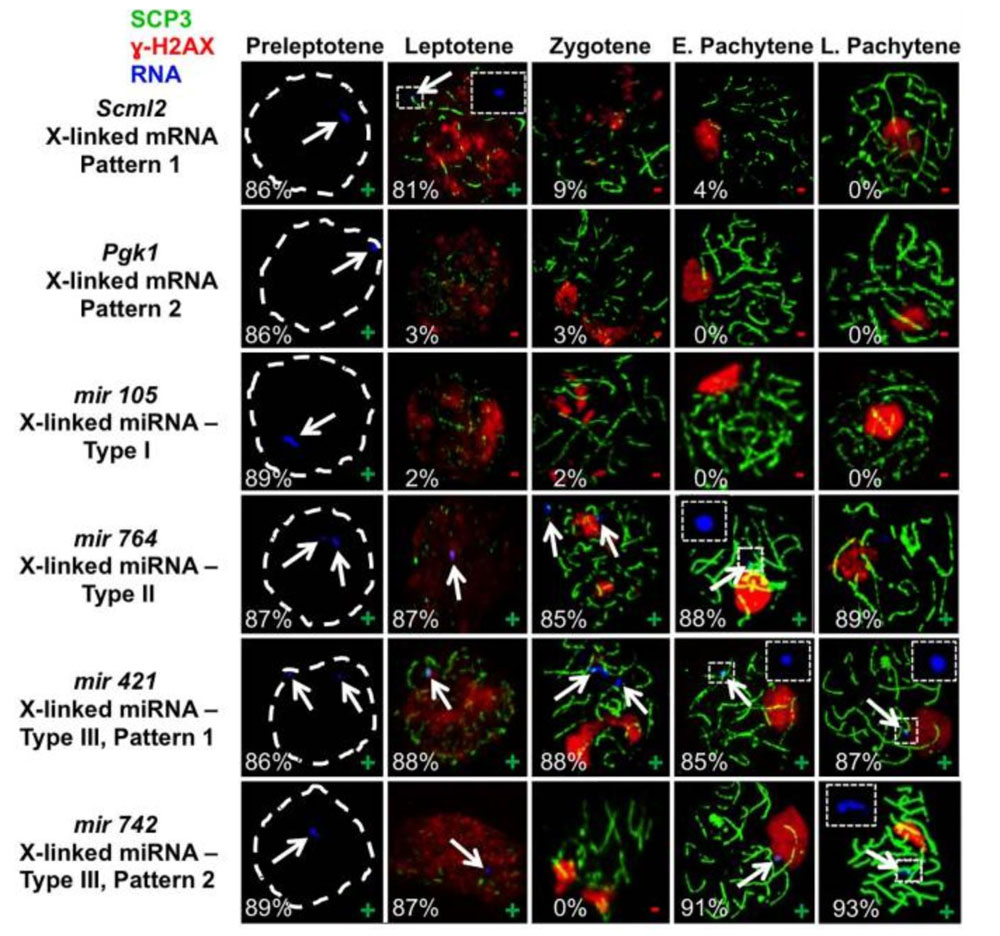 During meiosis in male mammals the X and Y sex chromosomes undergo a transient transcriptional inactivation in spermatocytes termed meiotic sex chromosome inactivation (MSCI). We previously showed that the Xist gene is expressed in spermatogenic cells but is not required to initiate MSCI. We then showed that MSCI occurs in both eutherian and metatherian mammals. Finally, in collaboration with Dr. Wei Yan we showed that while no X-linked mRNA-encoding genes have been shown to escape MSCI, many X-linked microRNA-encoding genes do escape MSCI.
During meiosis in male mammals the X and Y sex chromosomes undergo a transient transcriptional inactivation in spermatocytes termed meiotic sex chromosome inactivation (MSCI). We previously showed that the Xist gene is expressed in spermatogenic cells but is not required to initiate MSCI. We then showed that MSCI occurs in both eutherian and metatherian mammals. Finally, in collaboration with Dr. Wei Yan we showed that while no X-linked mRNA-encoding genes have been shown to escape MSCI, many X-linked microRNA-encoding genes do escape MSCI.
- Sosa E, Flores L, Yan W, McCarrey JR. (2015) Escape of X-linked miRNA genes from meiotic sex chromosome inactivation. Development 142: 3791-3800.
- Song R, Michaels J, Ro S, Zhang J, McCarrey JR, Yan W. (2009) Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet 41:488-493.
- Yan W and McCarrey JR. (2009) Sex chromosome inactivation in the male. Epigenetics 4:1-5.
- Hornecker J, Robinson ES, Samollow PB, VandeBerg JL, McCarrey JR. (2007) Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 45: 696-708.
- Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT. (2007) Sex-chromosome silencing in the marsupial male germline. Proc Natl Acad Sci USA. 104: 9730-9735.
- Namekawa SH, Park PJ, Zhang L-F, Shima JE, McCarrey JR, Griswold MD, Lee JT. (2006) Postmeiotic sex chromatin in the male germline of the mouse. Curr. Biol. 16: 660-667.
- Wang J, Page D, McCarrey JR. (2005) Differential expression of sex-linked and autosomal germ-cell specific genes during spermatogenesis in the mouse. Hum Molec Genet 14: 2911-2918.
- McCarrey, J.R., Watson, C., Atencio, J., Ostermeier, G.C., Marahrens, Y., Jaenisch, R., and Krawetz, S.A. (2002) X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis. 34: 257-266.
- McCarrey, J.R. (2001) X-chromosome inactivation during spermatogenesis: The original dosage compensation mechanism in mammals? In: Gene Families: Studies of DNA, RNA, Enzymes and Proteins G. Xue, Y. Xue, Z. Xu, R. Holmes, G. L. Hammond, H. A. Lim (eds). World Scientific Publishing Co. New Jersey. pp. 59-72.
- Wang, P.J., McCarrey, J.R., Yang, F. and Page, D.C. (2001) An abundance of X-linked genes expressed in spermatogonia. Nature Genetics. 27:422-426.
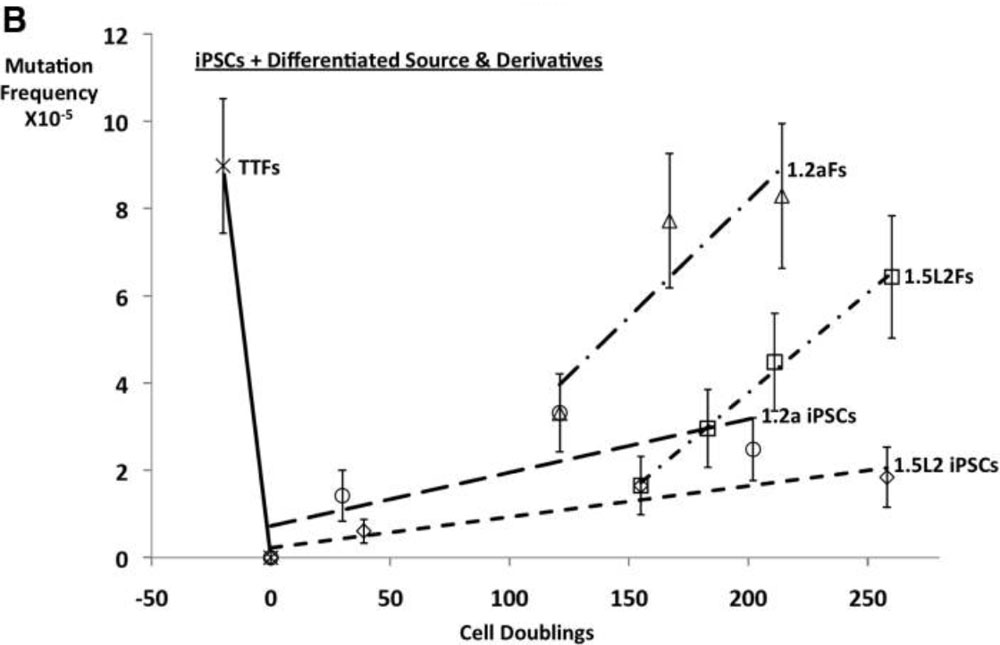 In collaboration with Dr. Christi Walter at UT Health San Antonio, we have shown that as predicted by the "Disposable Soma Theory," both germ cells and pluripotent cells maintain genetic integrity at elevated levels relative to somatic or differentiated cells, respectively, and that this is based on enhanced expression of DNA repair and cell death genes regulated in concert with genes driving pluripotency or germ cell fate.
In collaboration with Dr. Christi Walter at UT Health San Antonio, we have shown that as predicted by the "Disposable Soma Theory," both germ cells and pluripotent cells maintain genetic integrity at elevated levels relative to somatic or differentiated cells, respectively, and that this is based on enhanced expression of DNA repair and cell death genes regulated in concert with genes driving pluripotency or germ cell fate.
- Cooper D, Walter CA, McCarrey JR. (2017) Pluripotent Cells Display Enhanced Resistance to Mutagenesis. Stem Cell Research 19:113-117.
- Chen I-C, Hernandez C, Xu X, Cooney A, Wang Y, McCarrey JR. (2016) Dynamic variations in genetic integrity accompany changes in cell fate. Stem Cells Devel Nov 15;25(22):1698-1708.
- Cooper D, Walter CA, McCarrey JR. (2014) Co-regulation of pluripotency and enhanced maintenance of genetic integrity at the genomic level. Stem Cell Res 13: 508-519.
- Murphey P, McLean D, McMahan CA, Walter CA, McCarrey JR. (2013) Enhanced genetic integrity in mouse germ cells. Biol. Reprod. Jan 3;88(1):6. doi:10.1095/biolreprod.112.103481
- Walter, C.A., Intano, G.W., McCarrey, J.R., McMahan, C.A., and Walter, R.B. (1998) Mutation frequency declines during spermatogenesis in young mice but increases in old mice. Proc. Natl. Acad. Sci. (USA) 95: 10015-10019.
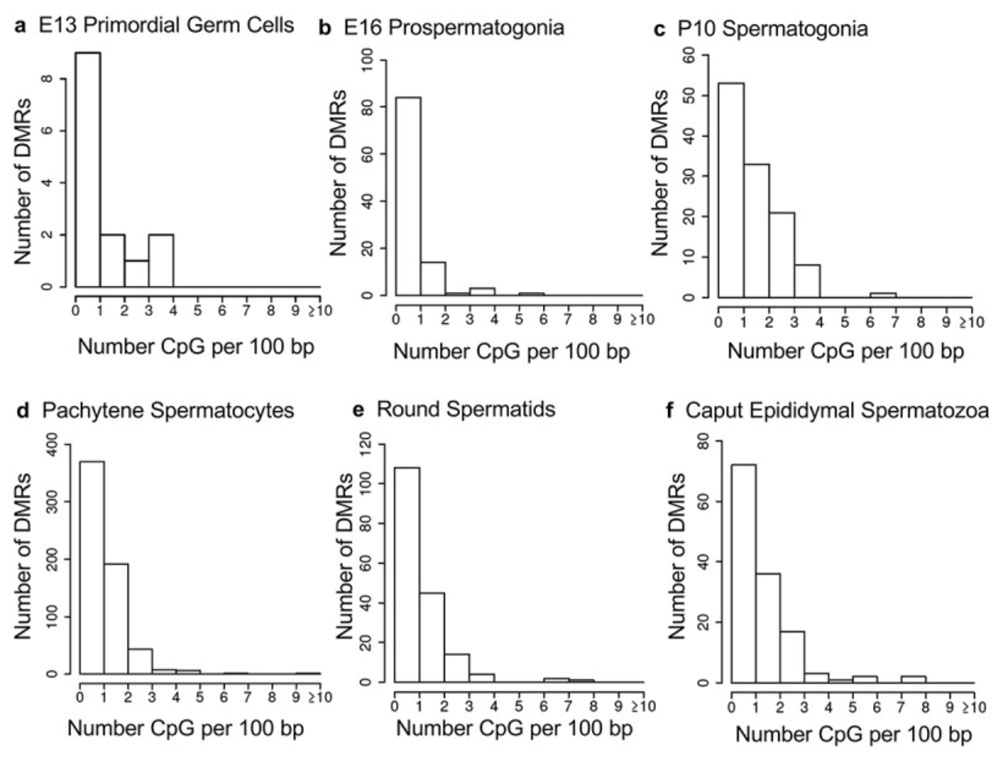 Disruptive environmental effects including exposure to certain chemicals, or aberrant dietary effects or stress or other conditions can induce epimutations that can then be transmitted from the exposed individual to his or her descendants over multiple generations. In collaboration with Dr. Michael Skinner at Washington State University, we have characterized this transgenerational epigenetic inheritance, especially with respect to how it is mediated by the germ line.
Disruptive environmental effects including exposure to certain chemicals, or aberrant dietary effects or stress or other conditions can induce epimutations that can then be transmitted from the exposed individual to his or her descendants over multiple generations. In collaboration with Dr. Michael Skinner at Washington State University, we have characterized this transgenerational epigenetic inheritance, especially with respect to how it is mediated by the germ line.
- Ben Maamar M, Beck D, Nilsson E, McCarrey JR, Skinner MK. (2019) Developmental origins of transgenerational sperm histone retention following ancestral exposures. Dev Biol. 2019 Jan 15;445(2):280-293. Epub 2018 Nov 27. doi:10.1016/j.ydbio.2018.11.016 | PMID:30500333
- Skinner MK, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR. (2019) Transgenerational sperm DNA methylation epimutation developmental origins following ancestral vinclozolin exposure. Epigenetics, 14:721-739. Epub 2019 May 13. doi:10.1080/15592294.2019.1614417 | PMID:31079544
- Maamar MB, Nilsson E, Sadler-Riggleman I, Beck D, McCarrey JR, Skinner MK. (2019) Developmental origins of transgenerational sperm DNA methylation epimutations following ancestral DDT exposure. Devel Biol, 445:280-293. Epub 2018 Nov 27. doi:10.1016/j.ydbio.2018.11.016 | PMID:30500333
- McCarrey JR, Lehle JD, Raju SS, Nilsson EE, Skinner MK. (2016) Tertiary epimutations – a novel paradigm contributing to epigenetic transgenerational inheritance. Plos One Dec 19;11(12):e0168038. doi:10.1371/journal.pone.0168038
- Skinner MK, Gurerrero-Bosagna C, Haque Md, Nilsson E, McCarrey JR. (2013) Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and the subsequent germ line. PloS One Jul 15;8(7): e66318.
- McCarrey JR, (2012) The epigenome as a target for heritable environmental disruptions of cellular function. Mol Cell Endocrinol 354: 9-15.

